National Grants
Project code: PN-III-P1-1.1-TE-2021-1161
Contract number: TE 154/2022
Project title: Blood circulating molecular chaperons - potential markers for gestational diabetes evolution; a proteomic analysis
Acronym: PRE-GDM
Project duration: 02/06/2022 - 31/05/2024
Project director: Dr. Viorel Suica
Abstract:
Gestational diabetes mellitus (GDM), described as glucose intolerance that is first diagnosed in pregnancy represents a hazard factor for long-term maternal and offspring cardiometabolic disease. Molecular chaperones play crucial parts in assisting polypeptide folding, chaperoning and cryoprotection inside the cells. However, novel information now associates these proteins as relevant participants in the extracellular surroundings, with involvement in vital stress-related mechanisms akin to inflammation and immunity. We hypothesize that a specific panel of chaperons could predict the onset of GDM. The project goal is to establish a panel of blood circulating chaperons that could quantitatively correlate with the pathogenesis and evolution of GDM. Our specific objectives are to set up of a dependent correlation between the expression pattern of circulating extracellular chaperones and GDM debut and to standardize a chaperone absolute quantification methodology that can be readily transferred to clinical practice. For this, we will use the “selected reaction monitoring” strategy, as a recognized technique for high-throughput, multiplexing analysis of proteins with a significantly increased value over the alternative antibody-based approaches. This technique assures the required sensitivity, selectivity and non-invasive measurement and will represent a cost-effective, valuable asset for the rapid transfer of basic biomedical research knowledge to clinical practice.
Objectives:
Objective 1: Correlation between the expression pattern of diabetic plasma circulating chaperones and GDM evolution;
Objective 2: Validation of the identified pattern of chaperones as potential markers of GDM evolution by an absolute quantitative profile of the chaperones.
The proposed workflow can be found below (Figure 1).
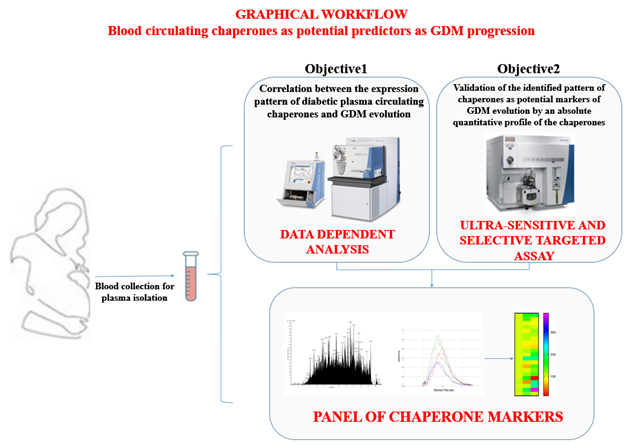
Figure 1: Graphical workflow of the project.
Report stage 2022:
Objective 1: Correlation between the expression pattern of diabetic plasma circulating chaperones and GDM evolution;
- Activity 1.1: Patients’ professional enrollment and plasma sample collection
- Activity 1.2: Plasma sample preparation and optimization for mass spectrometric analysis
- Activity 1.3: Mass spectrometric analysis for the identification of circulating chaperones
At this stage, in collaboration with the National Institute of Diabetes, Nutrition and Metabolic Diseases "N. Paulescu", the process of recruitment of patients with GDM, control subjects (CG), clinical and paraclinical examination and collection of blood samples for plasma isolation was initiated. In this regard, the necessary approvals for the collection of human samples have been obtained, through the completion of patient informed consent and ethics documentation. Human samples started to be collected in the third trimester of gestation for the proposed proteomic analyses.
GDM was diagnosed by the 75-g oral glucose tolerance test (GTT) two hours after ingestion of aqueous glucose solution, as recommended by the National Institute for Health and Care Excellence (NIHCE), American Diabetes Association (ADA). GDM was diagnosed if one or more of the glucose level values exceeded the cut-off: fasting ≥92 mg/dL, 1 h ≥180 mg/dL, 2 h ≥153 mg/dL. Patient exclusion criteria were based on age (40 years) and the presence of hypertension, nephropathy, preeclampsia, retinopathy, and psychiatric treatment.
To reduce the dynamic domain of blood proteins, enrichment of samples in extracellular exosomal vesicles was preferred. These were concentrated from 500 µL of sample collected from the two experimental groups. Exosome size and stability were determined by dynamic light scattering (DLS) on a Zetasizer Nano ZS (ZEN 3600). The exosome samples were suitably prepared for nano-chromatography tandem mass spectrometric analysis.
The Easy nLC II nano-chromatographic system was coupled to the LTQ Orbitrap Velos Pro hybrid mass spectrometer (Thermo Scientific, San Jose, CA, USA) for peptide separation and mass spectra acquisition. For each sample, 1 µg of peptide in technological triplicate was separated in a 15 cm × 75 μm.d., C18, 3 μm, 120 Å analytical column (Thermo Fisher Scientific, Rockford, IL, USA), and separation was performed using a gradient of solvent B (acetonitrile with 0.1% (v/v) formic acid) over solvent A (water with 0.1% (v/v) formic acid) for 90 min at a flow rate of 300 nL/min.
The chromatographic method was optimized for optimal elution of exosomal peptides by varying the acetonitrile gradient. Thus, using a 2-step gradient, starting from 2% acetonitrile to 30% acetonitrile for 65 minutes, followed by another step of increasing the acetonitrile concentration to 50% for another 25 minutes, an optimal elution resolution of both hydrophilic and hydrophobic peptides was obtained (Figure 2).
Figure 2: Chromatographic elution of a representative exosomal peptide sample.
We chose to operate the mass spectrometer in a top 15 "Data Dependent" configuration at a resolution power of 60000 for full scanning, in the 350-1700 m/z range. The chosen fragmentation mode was collision induced dissociation with helium molecules for the mass spectral acquisition of the ion fragments.
The raw data were processed for protein identification using the Sequest HT search algorithm within Proteome Discoverer 2.4 software (Thermo Fisher Scientific, Rockford, IL, USA). The Uniprot/SwissProt Homo sapiens reference proteome (SwissProt TaxID = 9606, v 2019-10-04) was used, with methionine oxidation set as dynamic modification and carbamidomethylation of cysteine as the modification, while a maximum of two missed cleavages was allowed. A reverse database search was performed for strict False Discovery Rate (FDR) settings of proteins and peptides (<5%). Label-free relative quantification was performed using the precursor ion quantifier node and was based on the intensity of unique peptide precursors present in 90% of replicate features.
The effect of GDM pathology was revealed by multivariate static analyses, such as Principal Component Analysis and Hierarchical Clustering Heat-Map Analysis, which demonstrated exosome proteome modification in the GDM group compared to CG (Figure 3).
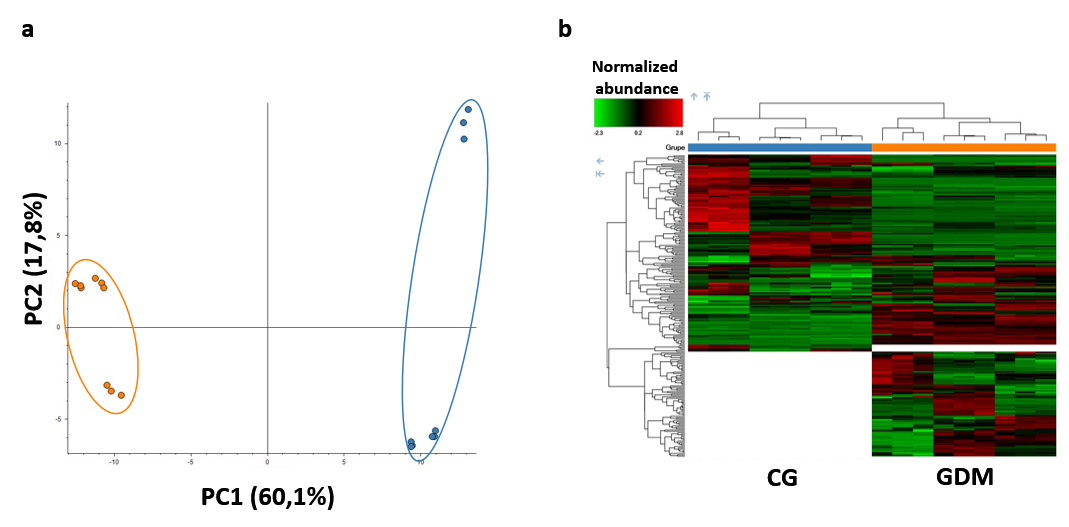
Figure 3: Multivariate statistical analyses: a) "Principal Component Analysis" and b) "Hierarchical Clustering Heat-Map Analysis" demonstrating an altered pathological state proteome (GDM) versus control (CG).
Report stage 2023:
Objective 1: Correlation between the expression pattern of diabetic plasma circulating chaperones and GDM evolution;
Objective 2: Validation of the identified pattern of chaperones as potential markers of GDM evolution by an absolute quantitative profile of the chaperones.
- Activity 2.1: Patients’ professional enrolment and plasma sample collection - continuation and completion;
- Activity 2.2: Plasma sample preparation and optimization for mass spectrometric analysis - continuation and completion;
- Activity 2.3: Mass spectrometric analysis for the identification of circulating chaperones - continuation and completion;
- Activity 2.4: Establishment of GDM relevant chaperones panel based on label-free quantification mass spectrometric experiments;
- Activity 2.5: Design and synthesis of proteotypic peptides (PTPs);
- Activity 2.6: Selection of the best ionizable peptides that uniquely identify the targeted proteins;
- Activity 2.7: Chromatographic elution and collision energy optimization;
- Activity 2.8: Selection of high intensity fragment ions for each specified PTP and chaperone molecule.
In this phase of the project, we focused on extracellular chaperone proteins in GDM vs. control samples by relative and then by absolute quantitative analysis. Firstly, the sample preparation process of the collected plasma was optimized by removing the 14 most abundant proteins. This step aimed at narrowing the dynamic range of existing protein concentrations and subsequently at identifying as many extracellular chaperone molecules as possible. By label-free mass spectrometric analysis, a pattern of chaperone molecules that are significantly regulated by gestational diabetes was obtained.
For these proteins, as well as for other extracellular chaperones, the in silico design process of proteotypic peptides was performed. The latter represent peptides that are unique to the targeted proteins with a special amino acid structure. By synthesizing stable isotopically labelled homologues, endogenous proteotypic peptides can be compared to the synthetic ones, which are injected into samples in well-defined amounts. Proteotypic peptides were selected using a complex bioinformatics analysis integrating machine learning algorithms. In silico design was performed for 27 proteotypic peptides associated with 13 circulating extracellular chaperone proteins.
Optimization steps have been initiated for Selected Reaction Monitoring analysis, an absolute quantification method using triple-quadrupole mass spectrometry. These include peptide ionization, chromatographic elution, peptide fragmentation, and selection of the most intense resulting ions.
Report stage 2024:
Objective 2: Validation of the identified pattern of chaperones as potential markers of GDM evolution by an absolute quantitative profile of the chaperones.
- Activity 3.1: Chromatographic elution and collision energy optimization;
- Activity 3.2: Selection of high intensity fragment ions for each specified PTP and chaperone molecule.
In this phase of the project, we focused on finalizing the optimization of some essential steps of the Selected Reaction Monitoring of the proteotypic peptides pertaining to the extracellular chaperon proteins.
We used several elution gradient formulas and time frames and demonstrated that although using a 10-min elution gradient, resulting in the co-elution of most of the 25 stable isotope-labeled peptides, the selectivity of the methodology allowed us to qualitatively and quantitively measure the peptides.
We optimized the collision energy (CE) necessary for the fragmentation of the precursor ions, using a 9-step normalized CE process.
Furthermore, using the best CE values obtained above, we selected the highest-intensity fragments for each peptide. These parameters prompted us to choose the best transitions for the quantification of the extracellular chaperons using SRM. We also performed standard curves with very good linearity response (R2>0.99) which will help us integrate the endogenous signals for determining their absolute quantity in the plasma of GDM patients.
Performance indicators:
ISI articles:
- Bernea EG, Suica VI, Uyy E, Cerveanu-Hogas A, Boteanu RM, Ivan L, Ceausu I, Mihai DA, Ionescu-Tîrgoviște C, Antohe F. Exosome Proteomics Reveals the Deregulation of Coagulation, Complement and Lipid Metabolism Proteins in Gestational Diabetes Mellitus. Molecules. 2022 Aug 26;27(17):5502. doi: 10.3390/molecules27175502
- Suica VI, Uyy E, Ivan L, Boteanu RM, Cerveanu-Hogas A, Hansen R, Antohe F. Cardiac Alarmins as Residual Risk Markers of Atherosclerosis under Hypolipidemic Therapy. Int J Mol Sci. 2022 Sep 22;23(19):11174. doi: 10.3390/ijms231911174
- Bernea EG, Uyy E, Mihai DA, Ceausu I, Ionescu-Tirgoviste C, Suica VI, Ivan L, Antohe F. New born macrosomia in gestational diabetes mellitus. Exp Ther Med. 2022 Oct 5;24(6):710. doi: 10.3892/etm.2022.11646
International manifestations:
- Oral presentations:
- Suica VI, Uyy E, Ivan L, Boteanu RM, Antohe F. Cardiac alarmins as residual risk markers of atherosclerosis under lipid-lowering therapy. 16th Central and Eastern European Proteomic Conference, 8th Informal Proteomic Meeting and 10th Czech Mass Spectrometry Conference, Praga, 2022.
- Suica VI, Uyy E, Ivan L, Boteanu RM, Uta DV, Antohe F. The critical role of proteomics in the application and effectiveness of experimental immuno-modulatory therapies. The 44th Annual Scientific Symposium of The Institute of Cellular Biology and Pathology "Nicolae Simionescu" held jointly with The 40th Annual Scientific Session of the Romanian Society for Cell Biology, Institutul de Biologie si Patologie Celulara "Nicolae Simionescu", București, 2023
- Posters:
- Ivan L, Uyy E, Suica VI, Boteanu RM, Antohe F. Residual hyperlipidemic stress under lipid lowering treatment may lead to irreversible NAFLD. 16th Central and Eastern European Proteomic Conference, 8th Informal Proteomic Meeting and 10th Czech Mass Spectrometry Conference, Praga, 2022.
- Uyy E, Suica VI, Boteanu RM, Ivan L, Antohe F. Arterial vessel wall proteome alteration involved in the regulation of cell death mechanisms in atherosclerosis. 16th Central and Eastern European Proteomic Conference, 8th Informal Proteomic Meeting and 10th Czech Mass Spectrometry Conference, Praga, 2022.
- Suica VI, Bernea G, Uyy E, Ivan L, Boteanu RM, Antohe F. Exosome proteomics reveals deregulated signaling pathways in gestational diabetes mellitusInternational Conference and XXXIX Scientific Session of the Romanian Society for Cell Biology, Cluj, 2022
- Uyy E, Suica VI, Ivan L, Antohe F. Non-apoptotic regulated cell death mechanisms under lipid-lowering therapy. International Conference and XXXIX Scientific Session of the Romanian Society for Cell Biology, Cluj, 2022
- Ivan L, Uyy E, Suica VI, Boteanu RM, Antohe F. Hepatic alarmins and mitochondrial dysfunction under residual hyperlipidemic stress led to irreversible nonalcoholic fatty liver disease. International Conference and XXXIX Scientific Session of the Romanian Society for Cell Biology, Cluj, 2022
- Ivan L, Suica VI, Uyy E, Uta DV, Antohe F. Functional proteomic changes of human monocytes in culture induced by hyperlipidemic stress. The 44th Annual Scientific Symposium of The Institute of Cellular Biology and Pathology "Nicolae Simionescu" held jointly with The 40th Annual Scientific Session of the Romanian Society for Cell Biology, Institutul de Biologie si Patologie Celulara "Nicolae Simionescu", București, 2023
- Uyy E, Suica VI, Ivan L, Uta DV, Bernea G, Georgescu DE, Ciriac O, Antohe F. Proteomic alterations in diabetic peripheral artery disease: insights from clinical and laboratory analyses. The 44th Annual Scientific Symposium of The Institute of Cellular Biology and Pathology "Nicolae Simionescu" held jointly with The 40th Annual Scientific Session of the Romanian Society for Cell Biology, Institutul de Biologie si Patologie Celulara "Nicolae Simionescu", București, 2023
- Uta DV, Suica VI, Uyy E, Ivan L, Antohe F. Differential proteomic analysis of lower limb ulcerations in an experimental peripheral disease diabetic mouse model. The 44th Annual Scientific Symposium of The Institute of Cellular Biology and Pathology "Nicolae Simionescu" held jointly with The 40th Annual Scientific Session of the Romanian Society for Cell Biology, Institutul de Biologie si Patologie Celulara "Nicolae Simionescu", București, 2023

